Radiogenomics, or imaging genomics, is a term that describes an exciting emerging field of research that is aimed at defining relationships between image features (image phenotypes) and molecular markers (molecular phenotypes).1 Traditionally, radiology has been primarily focused on the correlation between imaging and histopathologic findings.
Merging the fields of standard imaging and genetic methods is obviously not straightforward. Noninvasive brain imaging techniques are reliable to provide objective and reproducible phenotypes, are generally widely available, and in the case of brain tumor, remain the primary method for diagnosis, surgical guidance, and surveillance of tumors.
Medulloblastoma, a WHO grade IV embryonal tumor, is the most common malignant CNS tumor in children, accounting for 40% of childhood tumors in the posterior fossa.2 Approximately 338 new pediatric patients are diagnosed in the United States each year. As a result, tremendous efforts have been made toward its genomic classification.
The traditional classification system is based on histopathology, and according to the 2016 WHO Classification of Tumors of the Central Nervous System includes: classic medulloblastoma, desmoplastic/nodular medulloblastoma, medulloblastoma with extensive nodularity, and large cell/anaplastic medulloblastoma.3
Beyond histology, medulloblastoma is currently stratified into 4 genetic/molecular subtypes based on transcriptome profiling studies, as detailed in an article by Taylor et al.4 These subgroups include activated wingless (WNT), activated sonic hedgehog (SHH) with or without TP53 mutation, and non-SHH/non-WNT (group III and group IV tumors).3 Each subgroup is unique in its origin and pathogenesis, as well as its prognosis and potential therapeutic options. The SHH and WNT subgroups correlate with the site of origin of these tumors.5
This installment of the AJNR News Digest highlights several pediatric medulloblastoma studies that contribute to our understanding of MR imaging features of the tumor, including location, enhancement patterns, and MR spectral characteristics, which help to optimize our capacity to predict molecular subgroups of medulloblastoma. The first study that analyzes MR imaging biomarkers for genetic entity discrimination of adult medulloblastomas from pediatric medulloblastomas is highlighted.6 This edition also features a review of radiomics in brain tumor and a study aimed at improving current posterior fossa discrimination of tumor type by using support vector machine classifiers on quantitative MR imaging features.7,8
The potential to identify molecular subtypes noninvasively aids in prognostication and risk-adaptive treatment strategies. Targeted therapeutic intervention, escalation, or de-escalation of treatment therapy to include the avoidance/minimization of radiation, with the goal of improving outcomes for patients with group III and IV medulloblastomas and reducing late effects of toxic therapy, are important benefits to patients. Despite the potential utility of clinical genomic analyses, their translation into clinical practice to improve treatment outcomes can be hampered by cost or lack of access to molecular analysis tools when treatment is initiated.
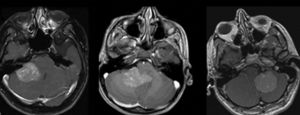
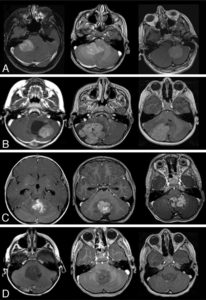
A study by Perreault et al2 demonstrated that a molecular subgroup could be correctly predicted by neuroimaging in 65% of 99 retrospectively reviewed medulloblastoma cases, based on tumor location and enhancement pattern. In terms of location, WNT pathway tumors were most likely to arise in the cerebellopontine angle cistern or cerebellar peduncle, whereas SHH pathway tumors were found in the cerebellar hemispheres. Group III and group IV tumors were the primary subgroups encountered in the midline fourth ventricle. Group III tumors tended to be ill-defined.
Unlike that observed in adults, the value of contrast enhancement is limited in predicting tumor grades in children, as 89% of low-grade tumors will show enhancement. Moreover, lack of enhancement is present in high-grade tumors such as primitive neuroectodermal tumors, and a significant percentage of medulloblastomas present minimal or no enhancement. In the study by Perreault et al,2 group 4 tumors were characterized by absent or minimal contrast enhancement. This is an important observation because group IV medulloblastomas are currently not well-differentiated by using immunohistochemistry markers.2
Patay et al5 further refines the spatial distribution of WNT subgroup medulloblastomas. The results of the study suggest that tumors are close to the midline but are lateralized, originating from the brain stem and cerebellum around the foramen of Luschka, which is in concordance with the current understanding of the embryologic origins of this tumor subtype.
Low ADC values are characteristic of medulloblastomas, reflecting the highly cellular nature of these tumors. Differences in ADC values have been used to distinguish cellular medulloblastomas from pilocytic astrocytomas. Distinctive MRI ADC has not yet been shown to correspond to the genetic subtype of medulloblastoma or treatment responsiveness.8
Medulloblastomas across molecular subgroups reveal distinct MR spectral features. Bluml et al9 showed that pediatric group III and group IV tumors demonstrate metabolic profiles with readily detectable taurine, lower levels of lipids, and high levels of creatine. SHH tumors showed prominent choline and lipid levels with low levels of creatine, and little or no evidence of taurine. A 5-metabolite subgroup classifier inclusive of creatine, myoinositol, taurine, aspartate, and lipid can distinguish between group III/IV and SHH medulloblastomas with excellent accuracy.
Keil et al6 identified MR imaging biomarkers in a cohort of 28 adult patients with medulloblastoma; these included a small tumor volume, fourth ventricular contact, and the absence of hydrocephalus in WNT tumors. No coherent MR imaging biomarkers for the same genetic entities could be identified between the adult cohort and corresponding data from a pediatric study, possibly due at least in part to the small size of the adult cohort.