Joseph Maldjian
Arterial spin-labeling (ASL) perfusion MRI has been in development for over a decade.1–3 During much of this time it had remained predominantly a research tool. I was a neuroradiology fellow at the University of Pennsylvania when pioneering work in this area was being conducted by Drs. Detre and Alsop. They were developing the continuous arterial spin-labeling (CASL) methodology and extending it to multisection imaging.2 Later, as a faculty member there, I was able to spend some time working with them as part of an ARRS scholar award.
Ever since learning about ASL during my fellowship, I have been interested in the clinical translation of this technique. Two major hurdles were hampering its clinical deployment. The first was development of robust pulse sequences for use in clinical populations with existing scanners. The second was automation of the postprocessing, with integration into the clinical PACS for seamless integration into the clinical workflow. With the advent of pulsed arterial spin-labeling (PASL)4,5 and, more recently, pseudocontinuous arterial spin-labeling (pCASL),6 the acquisition side of the equation became relatively stable. The major hurdle to clinical use remained implementation into daily workflow.
I had already been involved in developing automated processing pipelines for a wide variety of applications on the research side (including fMRI, voxel-based morphometry, ASL, DTI, etc), and on the clinical side for fMRI.7–9 Shortly after coming to Wake Forest in 2001, I implemented a fully automated fMRI clinical processing pipeline.7 It was a short step to modify it for use with the PASL (and now pCASL) images. Our initial experience on the clinical side revealed issues related to patient motion and scanner gradient stability that were not as apparent on the research side. After implementing an automated filtering method (essentially detecting and discarding bad image pairs), the processing became sufficiently robust for widespread clinical implementation at our site.10 Our automated processing pipeline made it possible to perform many thousands of cases with seamless integration into the workflow.8,10–20
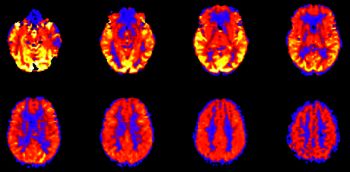
This paper in the AJNR was part of a 3-part series representing our initial experience with the technique in a very large number of cases.11–13 Some of the more important points include the appropriate identification of artifacts and a short differential for global hyperperfusion. ASL remains an integral part of our clinical work-up of a variety of pathologies, including brain tumors, ischemia, and seizure evaluation. We have reduced our pCASL acquisition time to approximately 2 minutes. More recently, we have been performing territory mapping in which the arterial supplying vessel can be visualized based on its color, and we plan to perform comparisons with other perfusion methods (including DSC and dynamic contrast-enhanced). ASL has also become a standard part of our research protocols. For example, our own recent work integrating biomechanics, MRI, MEG, and cognitive evaluations in youth and high school football players makes use of ASL as part of our multimodal MR protocol. This particular dataset represents the largest of its type in the world, and the acquisition of embedded helmet sensor data at all practices and games provides an extremely thorough characterization of the biomechanical forces experienced by the brains of young athletes.21,22
Using ASL and other advanced imaging techniques (eg, DTI, fMRI, SWI), we hope to determine the effects of sports-related subconcussive impacts on the brain and help make football a safer activity for millions of children.